PLUGINS#
Plugins allow for creating client-side custom visualizations, independent from the core functionalities of MINERVA. After clicking on the plugin icon a popup window becomes available (see images below).
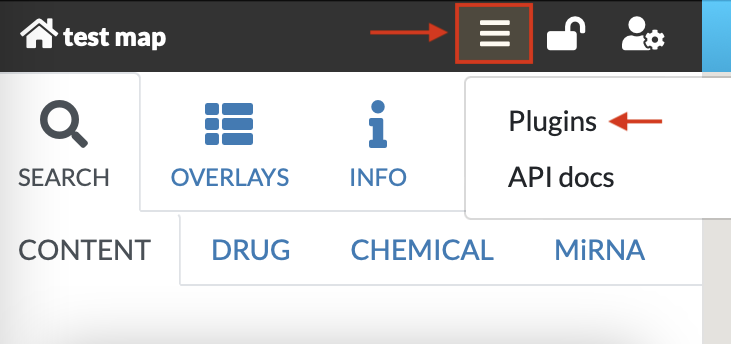
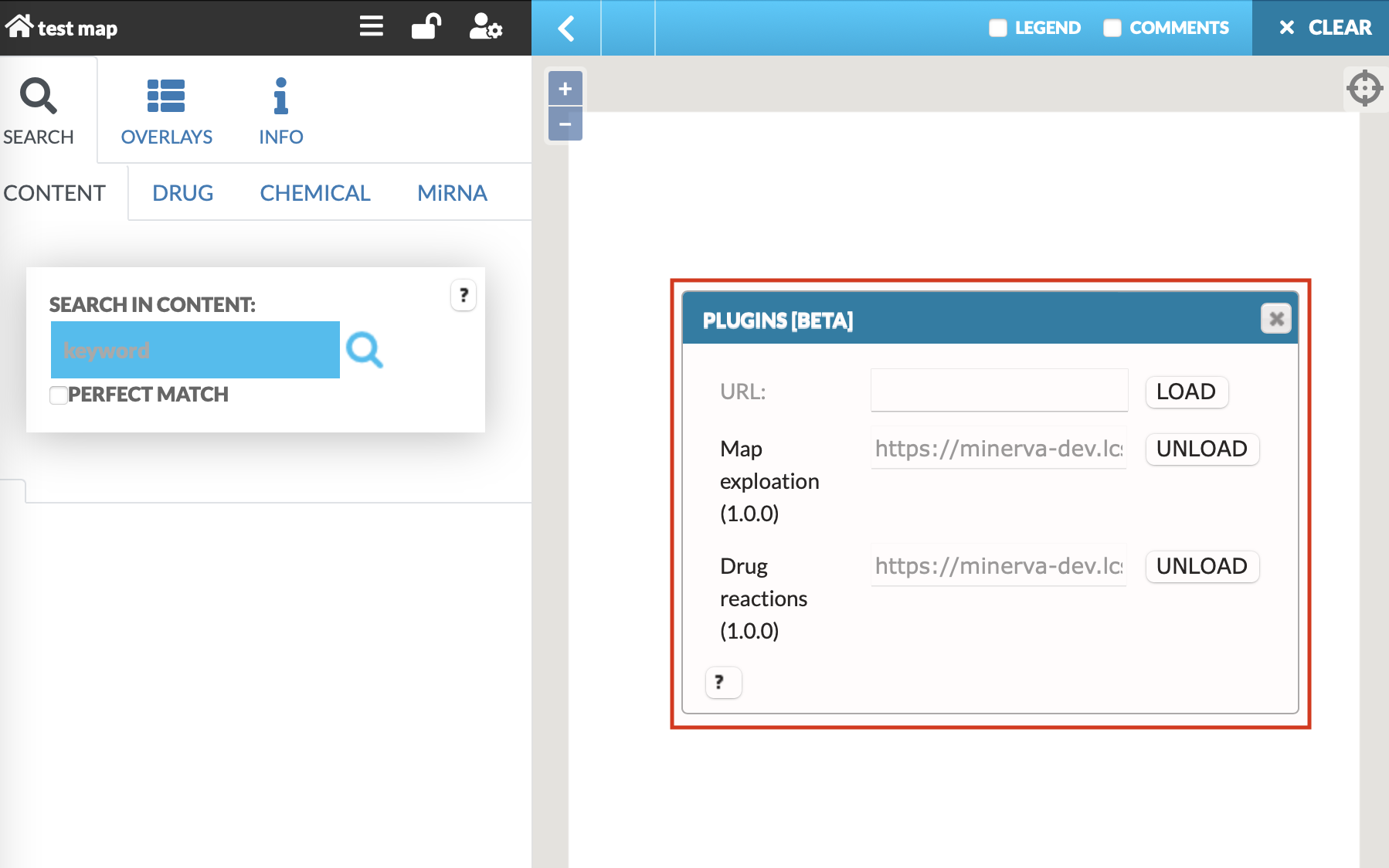
The user can choose any of the plugins that are available on the MINERVA platform, uploaded on the instance by the admin. Also, in the URL field one can supply a URL to the plugin, written in JavaScript. To learn how to write your own plugins, visit the “Plugin starter kit” repository.
Example plugin: you can upload below URL into the plugin dialog to find out more:
https://raw.githubusercontent.com/davidhoksza/minerva-plugins-starter-kit/master/dist/plugin.js
Afterwards, a plugin panel becomes available on the right (see image below). To close that panel, click the plugin icon to open the window and click Unload button.
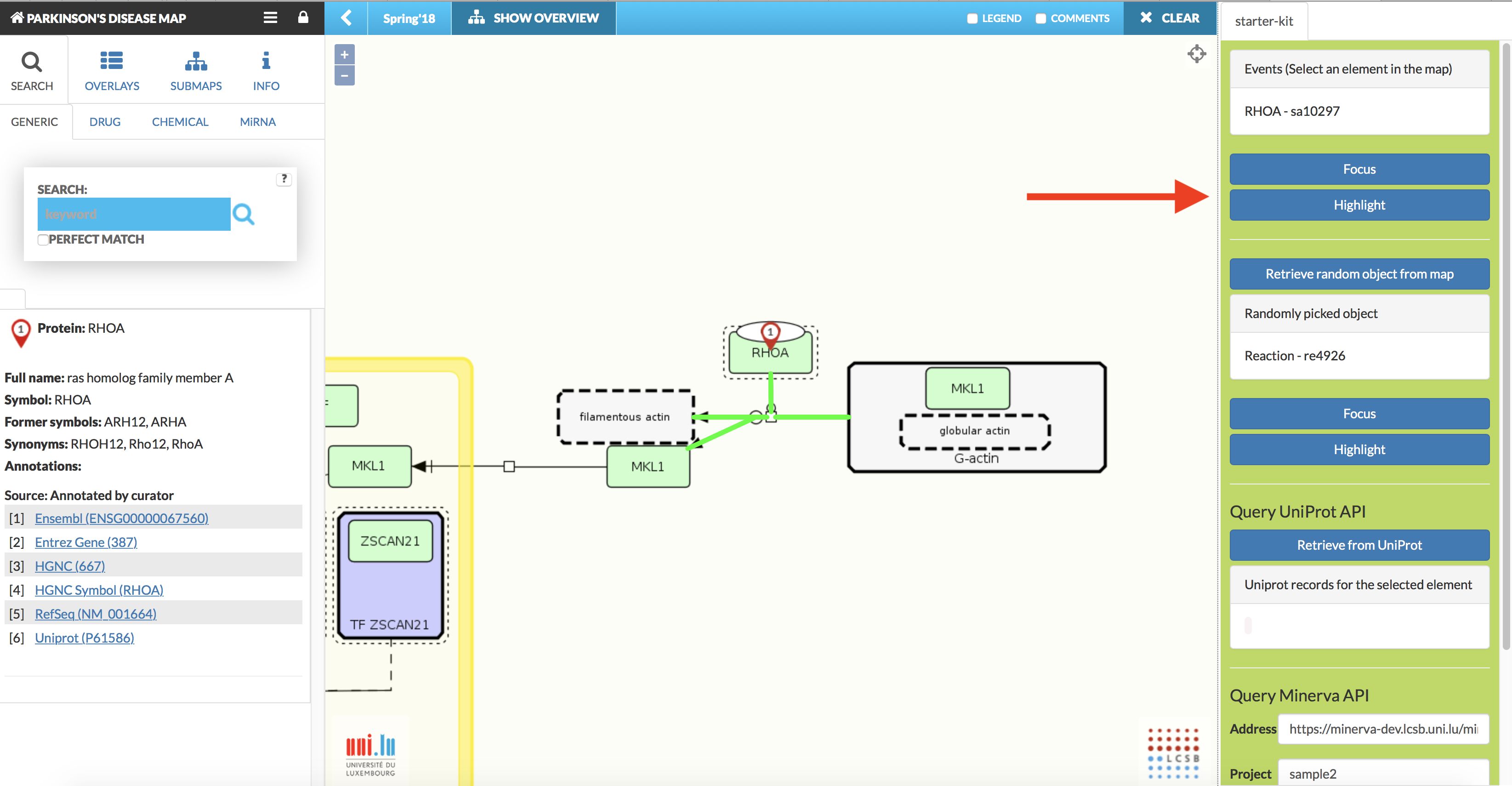
Please find below the list of available plugins, click at plugin’s name for its details.
| Name of the plugin | Address URL and the short description of the plugin |
|---|---|
| Disease variants associations | Provide the information about genes associated with the given disease in the context of the selected disease map. https://minerva-dev.lcsb.uni.lu/plugins/disease-associations/plugin.js |
| Drug reactions | Explore the adverse reactions of the drugs which are interacting with the presented entities in a disease map. https://minerva-dev.lcsb.uni.lu/plugins/drug-reactions/plugin.js |
| Exploration | Enhance the visualization and exploration of molecular interaction in the map. https://minerva-service.lcsb.uni.lu/plugins/exploration/plugin.js |
| Guide | Provide the introductory guide through various aspects of the MINERVA platform. https://minerva-dev.lcsb.uni.lu/plugins/guide/plugin.js |
| GSEA | Calculate the enrichment of the elements from the selected overlay in the pathway. https://minerva-service.lcsb.uni.lu/plugins/gsea/plugin.js |
–>