MINERVA platform: User’s manual#
User view refers to the visualization and functionalities accessed by users of the content hosted by your MINERVA instance.
You can also see the brief summary of this page in the MINERVA Quick Reference Guide or you can follow the guide.
MAIN VIEW#
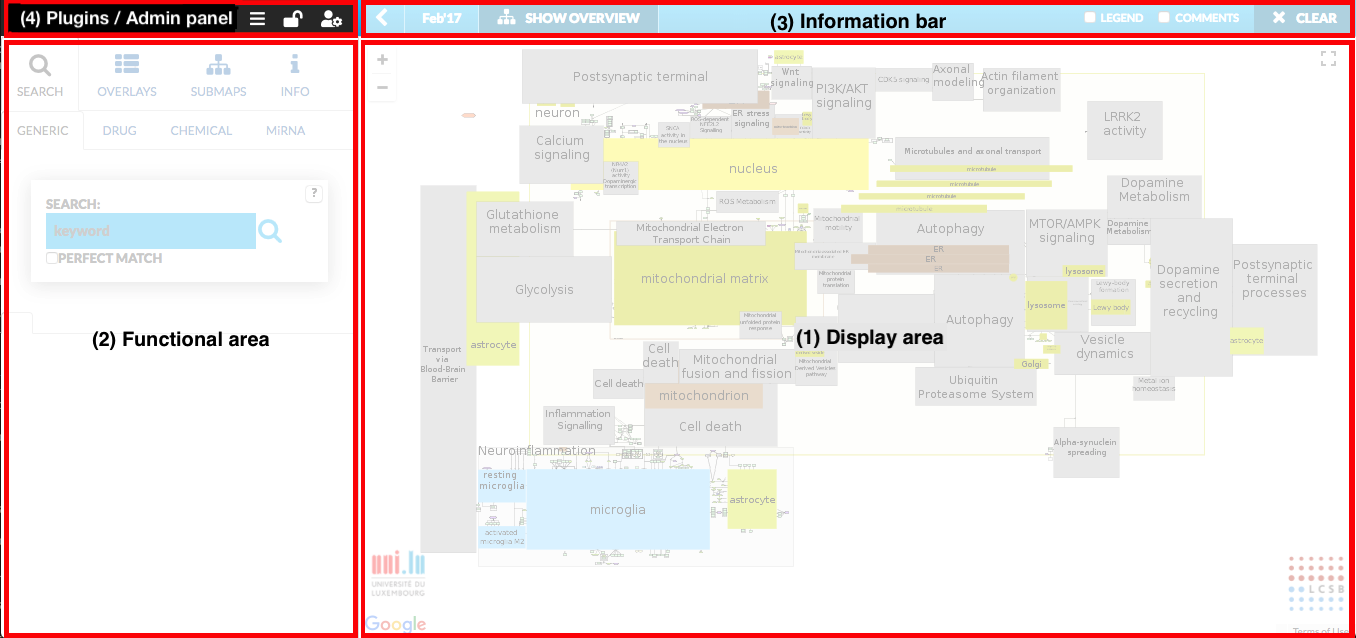
The main view of the MINERVA platform is summarized in the figure below.
 {:width=“900px”}
{:width=“900px”}
Main components of this view are:
- (1) Display area where the contents of the projects are visualized.
- (2) Functional area (called left panel), allowing for advanced interaction with explored content.
- (3) Information bar containing additional overlay functions.
- (4) Plugins / login-logout / administrator panel allowing a user on upload of own plugin, log in/out to MINERVA and enter Administrator panel. Login to MINERVA is possible also in Overlays tab or Info tab.
Accessing the project#
Your instance of MINERVA platform can host many projects, which are uploaded by the administrators (see add project). Every project project can be accessed by a dedicated web address.
Exploring the display area#
MINERVA platform displays molecular interaction diagrams, their annotations and links to databases.
Systems biology diagrams#
MINERVA platform visualises diagrams using three formats: SBGN-compliant, SBML and GPML. Read more about customising the content here.
The content is visualized using Google Maps API or Open Layers Library (chosen on the project upload), and allows for similar pan-and-zoom functionalities. The content is interactive, the user can click on an element or an interaction to examine additional details, which are displayed in the left panel (functional area). More exploration tools are available via right click on the map. Moreover, MINERVA allows to color the map’s elements with glyphs provided by user (check section glyphs) and to precisely set the elements order (check section Z-Index). The map content is by default displayed in semantic zoom mode, which enhances map’s clarity. In that view, the compartments and the pathways areas are displayed in a particular order, regulated by the zoom level.
Semantic zoom is a default view of the map. It makes the compartments and pathways cover underlying elements on higher levels of zoom. This view is generated procedurally, directly from the uploaded content. MINERVA calculates which areas cover others, and assigns them to the appropriate zoom levels. Note: In the tab Overlays tab in the left panel (functional area), semantic zoom is activated via Pathways and compartments overlay. You can turn it off by changing the view on Network or Empty.
Find out more here: procedural generation of the semantic zoom.
Basic exploration tools#
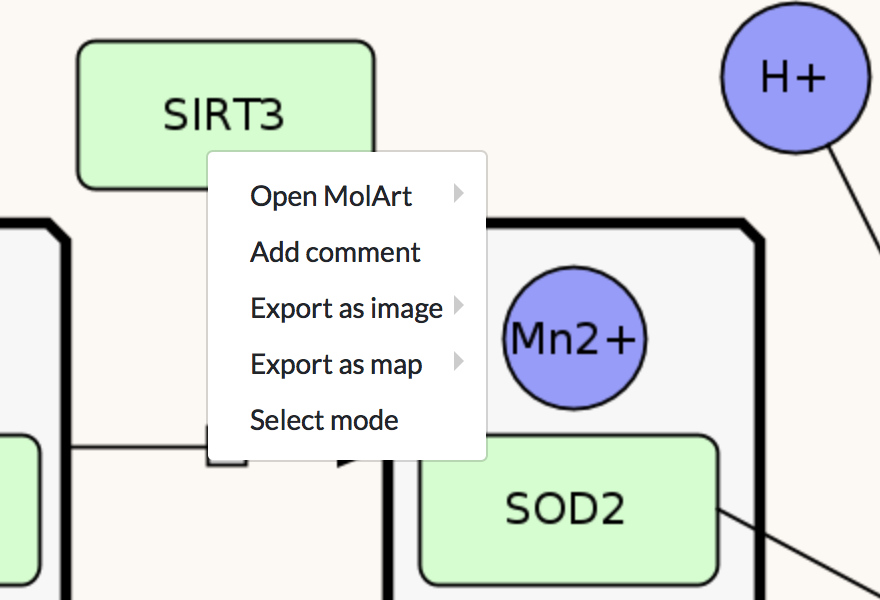
Right-click in the display area invokes a contextual menu (image below). The menu allows to:
- visualise the structures of proteins (MolArt),
- add a comment on a map,
- download map’s displayed area or particularly selected area (as image or map).
 {:width=“400px”}
{:width=“400px”}
Add comment#
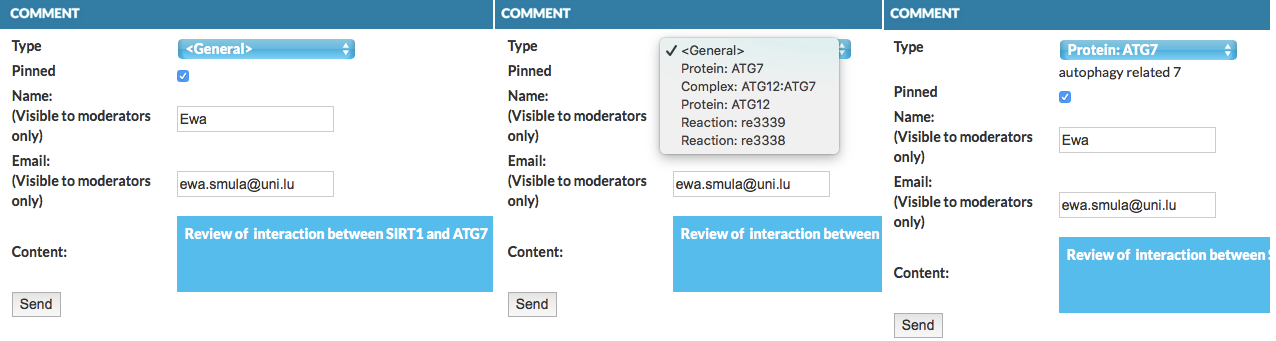
This functionality allows to comment on contents in the display area. Clicking on Add comment invokes a form that can be filled out by the user and sent to the administrators of the project (see below left). Type field is a drop-down menu, listing elements nearby to the click location. This allows to choose and attach the comment to a particular element or interaction (see below middle). If particular type is selected, its name is displayed (see below right). The field Pinned controls, whether the comment will be visible in the map.
 {:width=“900px”}
{:width=“900px”}
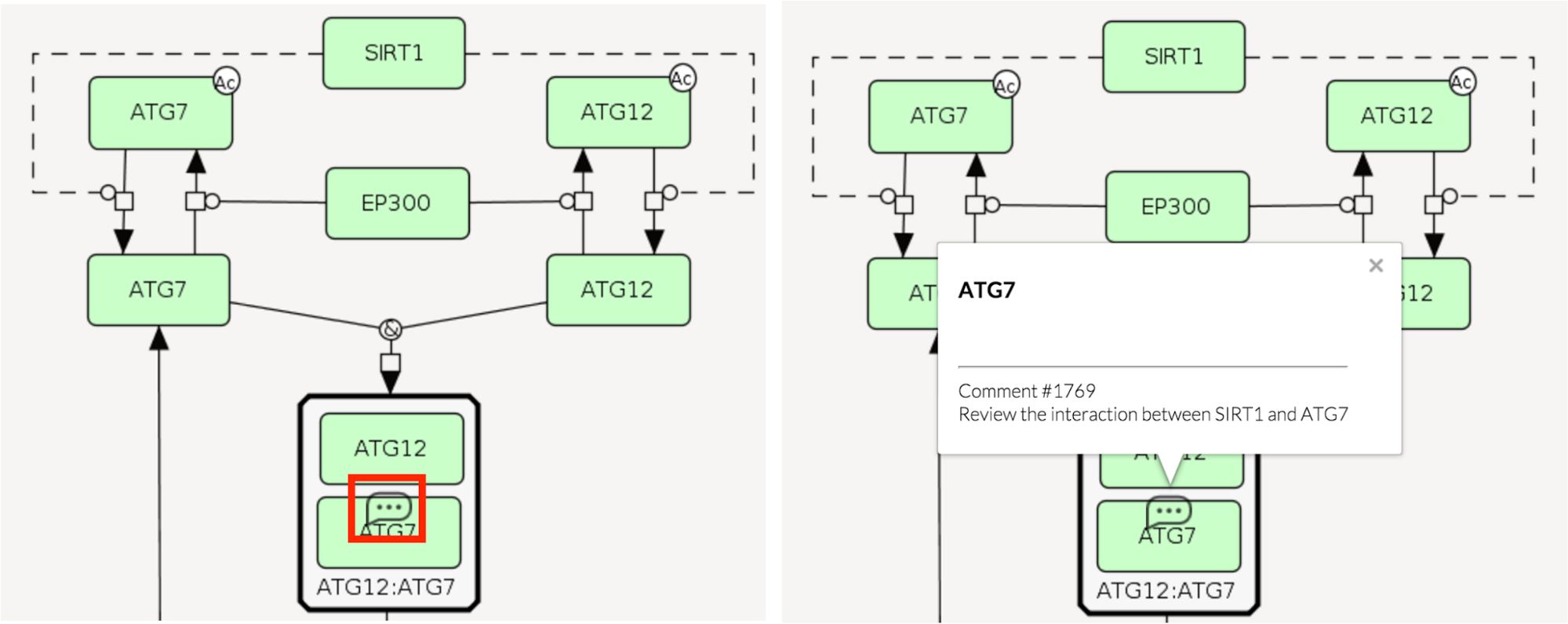
After sending, if the Pinned box was ticked, the comment bubble becomes visible in the display area (see below left). If the comment is placed in a submap, the main map shows that bubble also. It is placed on the anchor element. Note that the checkbox Comments located in Information bar must be ticked to make the comments visible at all.
 {:width=“900px”}
{:width=“900px”}
Clicking on the comment bubble displays annotated element and the text of the comment (see above right). Name and email are not disclosed, they will be accessible only to project and platform administrators (see Administrator’s manual - Comments).
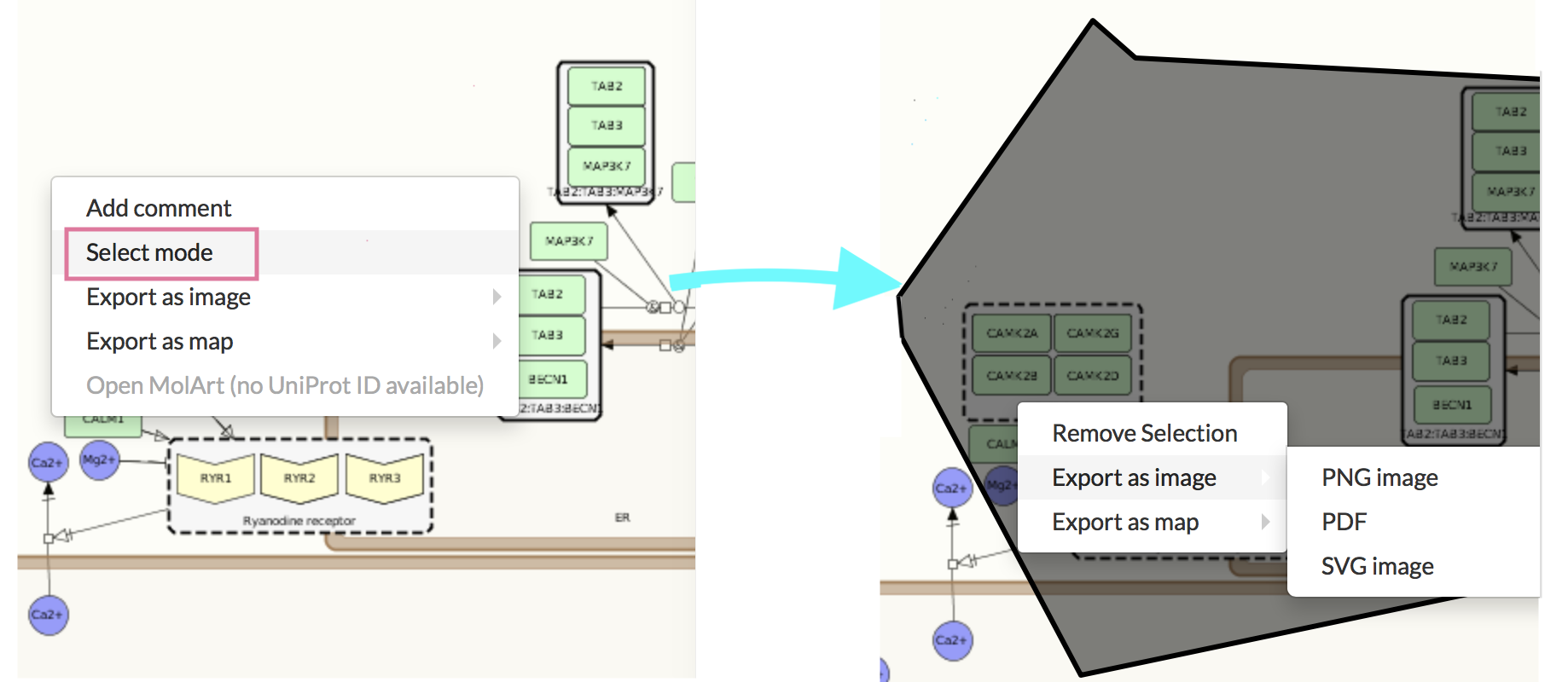
Select mode#
Clicking the Select mode enables the functionality supported by Google Maps API and Open Layers Library. It allows to select an arbitrary shape in the display area. To select the area: choose Select mode and click on the map, it is your start point. Draw all of your polygon’s points. To close the polygon choose the end point of the polygon the same as the start point, or double click on the end point. Right click on the selected area allows to remove the selection or download it in various formats, either as a map or an image.
 {:width=“900px”}
{:width=“900px”}
Export as image produces a rectangular image containing the selection. Export to vector and raster graphics is supported, allowing to modify and enhance the downloaded image.
Export as map produces a file acceptable by either CellDesigner, pure SBGN or SBML editors, containing the editable fragment of the selected display area, together with its contents and layout. Importantly, in the case of CellDesigner or SBML export, all additional annotations provided by MINERVA platform are downloaded as well.
Export as image#
This option creates an image of the part of map on your display area. Export to vector and raster graphics is supported, allowing to modify and enhance the downloaded image.
Export as map#
This option allows to produces a file acceptable by either CellDesigner, pure SBGN or SBML editors. The exported file contains the editable fragment of your display area, together with its contents and layout. Importantly, in the case of CellDesigner or SBML export, all additional annotations provided by MINERVA platform are downloaded as well.
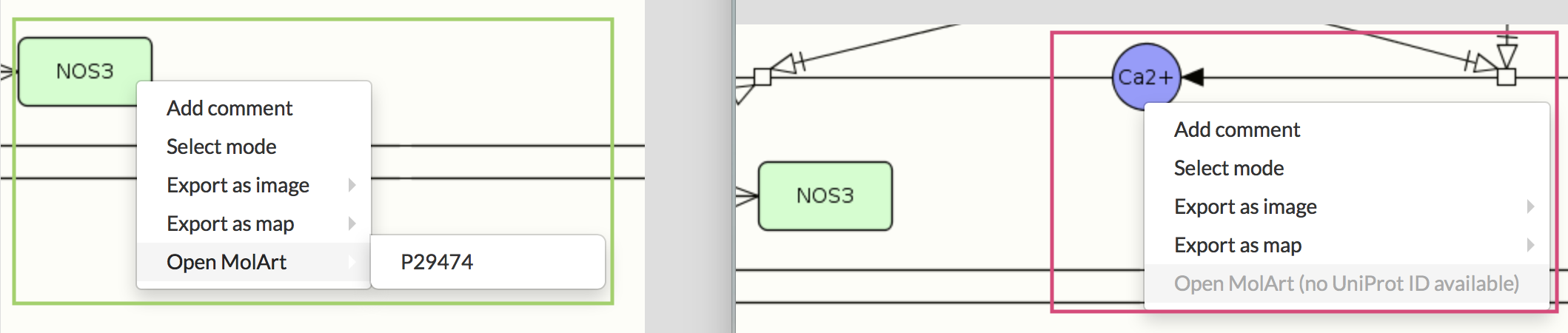
Open MolArt#
This option is active only for the protein annotated with UniProt ID (see image below). MolArt (MOLeculAR structure annoTator) is a JavaScript tool and library. It visualizes sequence-related annotations over available experimental or predicted structures (click here for MolArt documentation).
 {:width=“900px”}
{:width=“900px”}
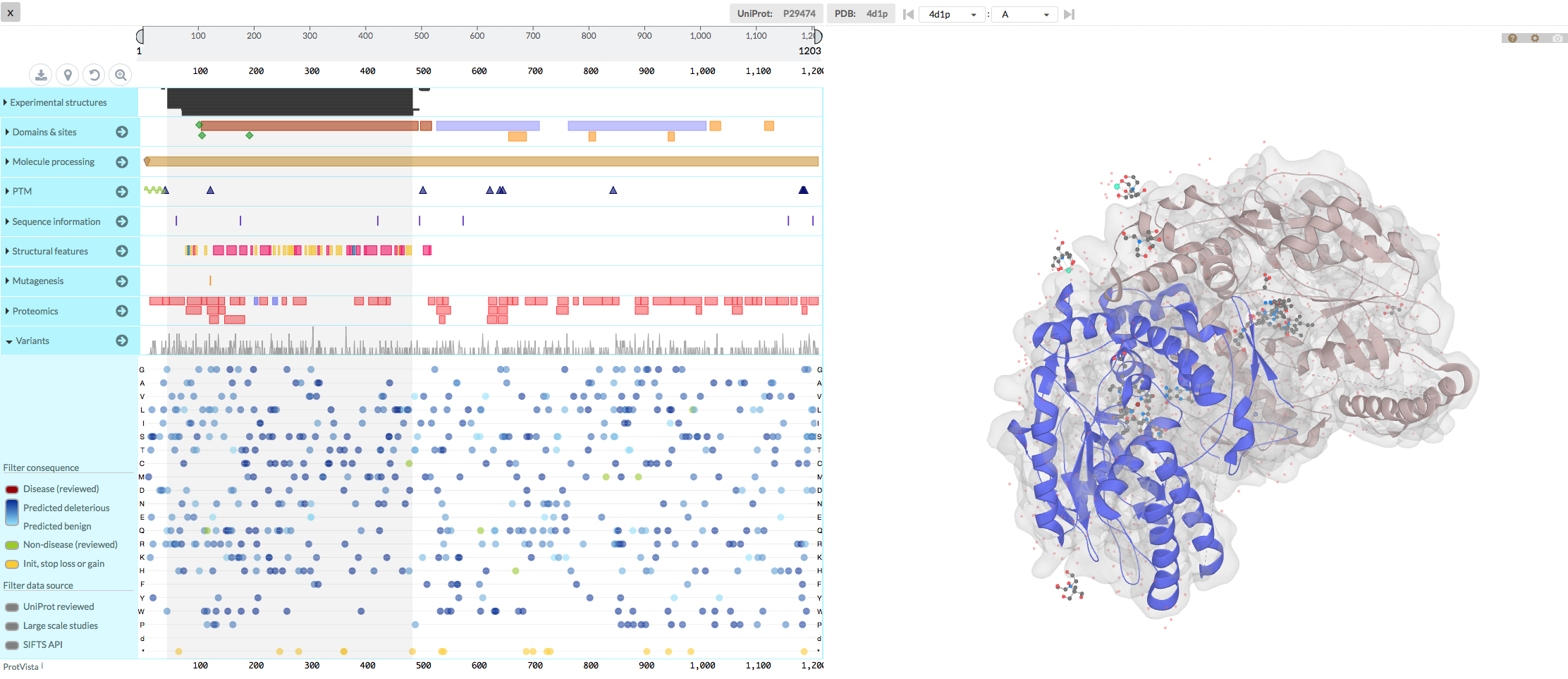
MolArt opens a new window, which displays in the left panel the molecule’s sequence, relevant annotations (including variation data) and the list of available structures (either experimental or predicted) for a given molecule. The right panel of MolArt window shows selected 3D structure over which any of the sequence annotations can be color-overlaid. See image below:
 {:width=“900px”}
{:width=“900px”}
FUNCTIONAL AREA#
Functional area (the panel to the left from the display area) displays additional information about selected elements and interactions, allows to query the content, generate custom overlays and browse the submaps.
Search tab#
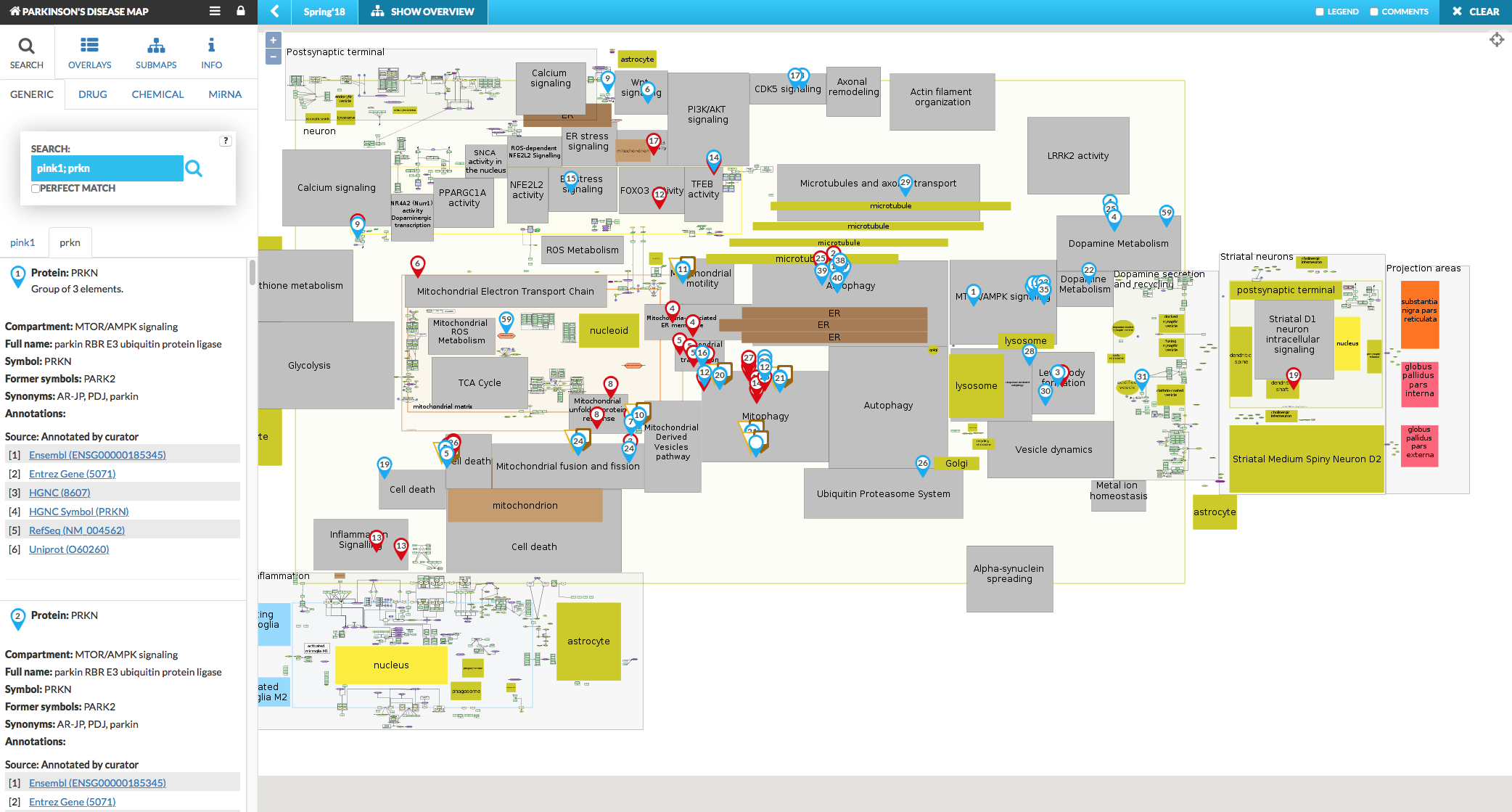
Search tab allows to search for a particular elements or interactions in the displayed map. Also, under this tab, the panel displays the detailed information on selected elements or interactions, such as: element’s compartment and full name, symbols, annotations and their source (automatic or manual by curator), see below:
 {:width=“900px”}
{:width=“900px”}
There are four subtabs available in the Search tab, offering different scopes:
- GENERIC for regular search
- DRUG for drug target search
- CHEMICAL for chemical target search
- MiRNA for miRNA target search
GENERIC#
The generic scope works as follows:
- Search field: Type your search query here, separating multiple elements with a semicolon ‘;’. Searches for separate terms will be visible in separate tabs. Search will look for similar names and synonyms of elements in the map. You can also search for an identifier of a reaction (visible in the left panel after clicking on a particular reaction). To do this, you need to add reaction: prefix to the searched term, e.g. for a reaction with id re944 you will search for a term reaction:re944. The + button shows/hides particular information as per Search panel options setting in MINERVA configuration.
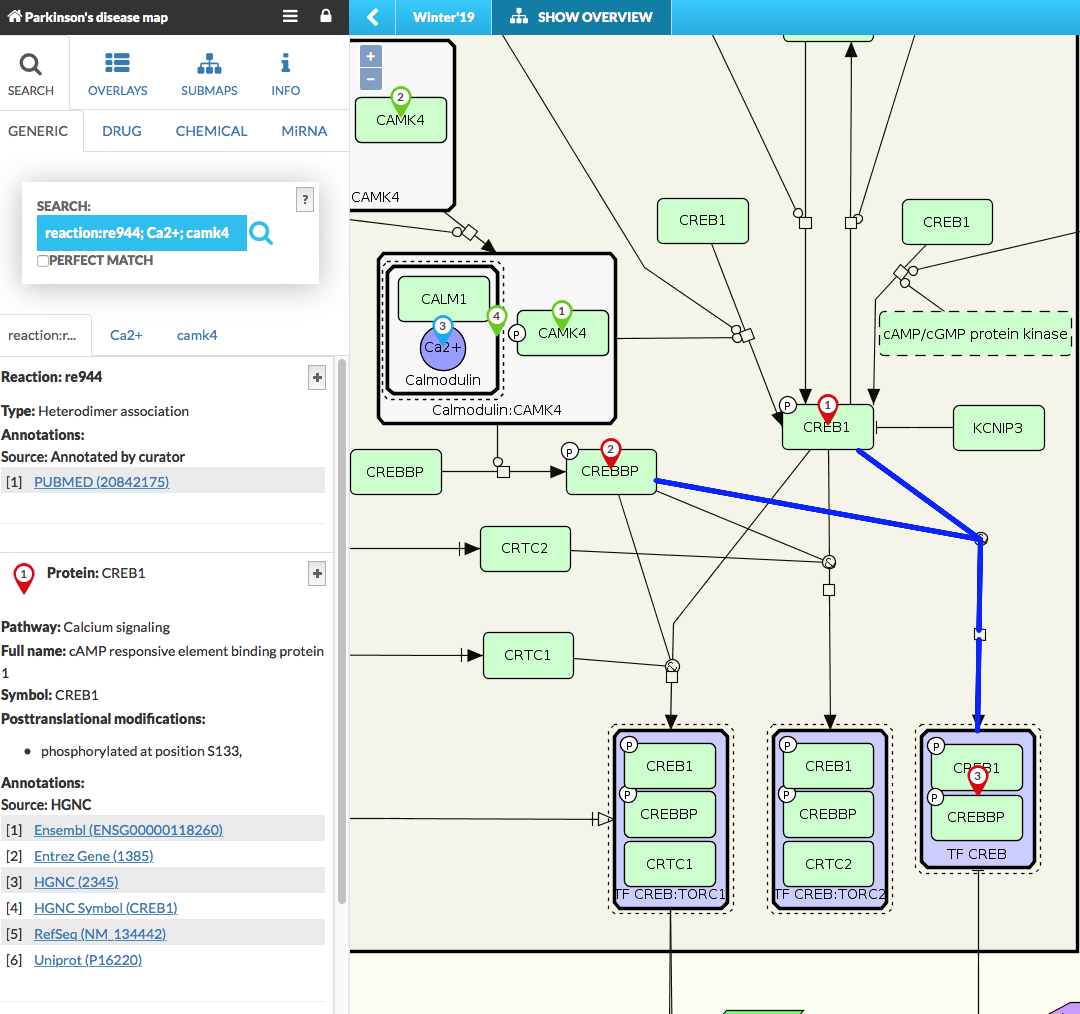 {:width=“900px”}
{:width=“900px”}
The reaction kinetics, if available, is displayed after clicking on chosen reaction in the display area. Information about element’s interactions is available for any element by clicking on its pin, see image below:
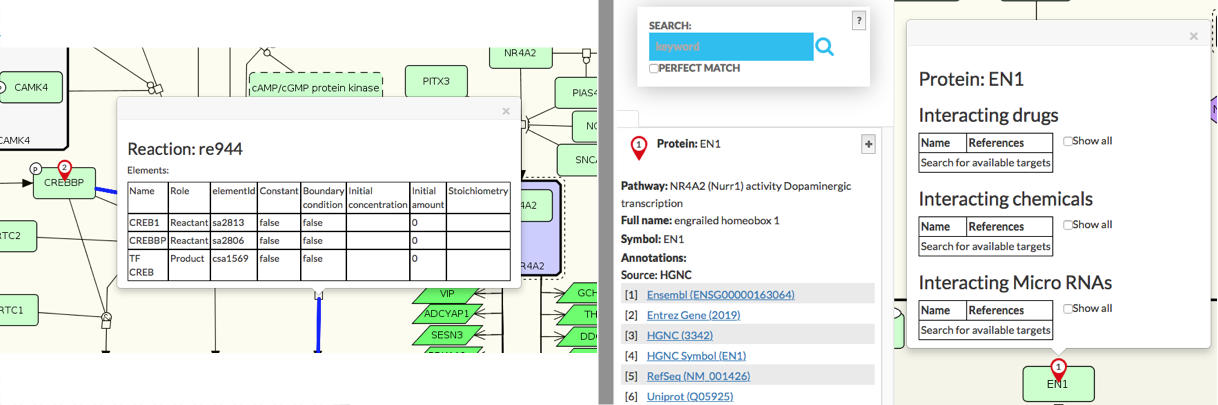 {:width=“900px”}
{:width=“900px”}
Search pins indicate hits in the display area. Similar search results are merged (marked with the same number) and the left panel contains information about size of that group of x elements. If search pin is be blank (no number), it would mean that aggregated search results exceed 100. For multiple elements search, the results have icons of different color and shape (see image below) which are clickable and show information in the popup window. When searching for different targets (e.g. generic target and drug target) common targets are highlighted with a special icon. The same icon indicates search results that are found in a submap. See more in Submaps tab.
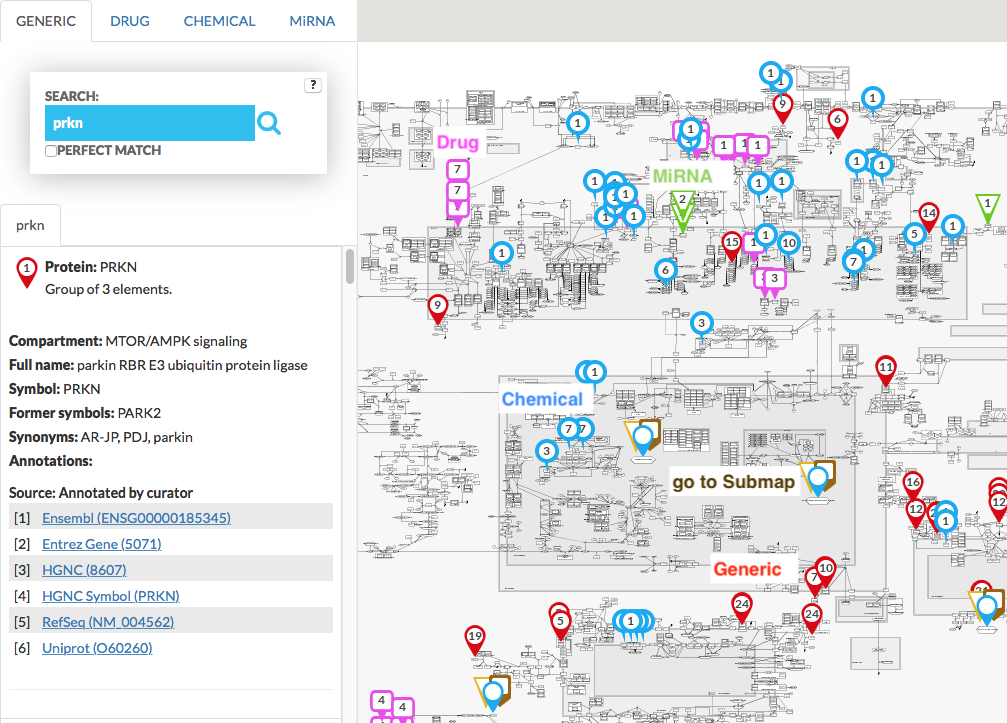 {:width=“900px”}
{:width=“900px”}
- Perfect match tick box: If this box is ticked, only terms exactly matching the query will be returned. For large networks or broad queries the search results may be capped to ensure the performance of the system. The limit of displaying search results can be configured by the administrator (see Administrator’s manual - Configuration).
Direct link to the elements in the display area is possible, as the search query can be provided within the web address of the displayed project. An address has to be manually constructed, directly into web browser, as below:
your.webserver.address/?search='search query'is a link executing the search for a given search query in the default project of your MINERVA instance. For example, an address
your.webserver.address/?search=reaction:xyz123will directly point to an interaction with id xyz123. Referring to a given project on your MINERVA instance requires additionally to pass the project identifier in the address, as shown below. Identifiers of projects are accessible using the Admin view.
your.webserver.address/?id='project id'&search='search query'DRUG (target)#
The Drug scope works as follows. Drug subtab under the the Search tab allows to look for known drugs or their brand names and display their targets in the map. The pins have specific shape, different than the results of other search scopes.
- Search field: type your search query here, separating multiple drug names or their brand names with a semicolon ‘;’. DrugBank (click) and ChEMBL (click) will be queried for known targets to be displayed in the map.
- Drug description, synonyms and all known targets will be displayed in the left panel.
CHEMICAL (target)#
The Chemical scope works as follows. Chemical subtab under the the Search tab allows to look for known chemicals or their synonyms and display their targets in the map. The pins have specific shape, different than the results of other search scopes.
- Search field: type your search query here, separating multiple chemical names or their synonyms with a semicolon ‘;’. Clinical Toxicogenomics Database (click) will be queried for known targets to be displayed in the map.
- Drug description, synonyms and all known targets will be displayed in the left panel.
MiRNA (target)#
The microRNA scope works as follows. MiRNA subtab under the the Search tab allows to look for miRNAs and display their targets in the map. The pins have specific shape, different than the results of other search scopes.
- Search field: type your search query here, separating multiple miRNAs with a semicolon ‘;’. miRTarBase (click) will be queried for known targets to be displayed in the map. Use only mature sequence IDs according to miRBase (click) (e.g. hsa-miR-125a-3p)
- Known targets will be displayed in the left panel.
Overlays tab#
Overlays tab allows to display or generate custom coloring of elements and interactions in the map. It is composed of two sections - general overlays and user-provided overlays (which becomes visible upon login).
General Overlays#
The table located in the top of the left panel (see image below), it contains:
- View column allows to change the viewing mode and display default multiple overlays:
- Viewing mode is changed by clicking on the magnifying glass icon, allows to switch between:
- Pathways and compartments (hierarchically masked content)
- Network (only colored network structure shown)
- Empty (only network structure, no coloring, recommended for data display)
- Display multiple overlays (tick chosen boxes) - choosing that option automatically turns the viewing mode to Empty to emphasize the overlay’s colors.
- Viewing mode is changed by clicking on the magnifying glass icon, allows to switch between:
- Data column contains buttons, where applicable, allowing to download the dataset used to generate the overlay.
Overlays provide additional information about elements and interactions. Hover over the overlay name/view/data to display a short description of the dataset, if provided on upload. Overlays are also visible in the popup window, displayed after clicking on the element’s pin, showing the value and the sign marked on the horizontal color bars. The popup contains information about Interacting drugs, chemicals and MicroRNA with selected element, available after ticking check box Show all.
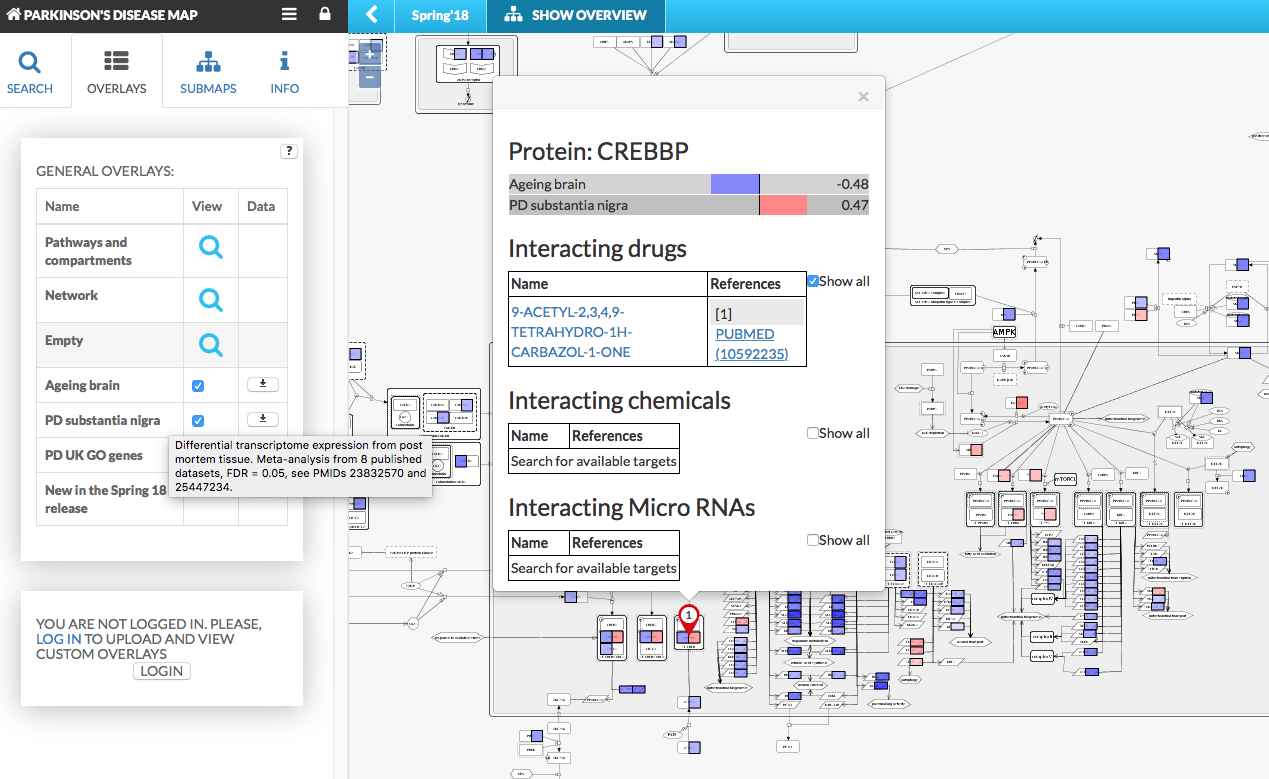 {:width=“900px”}
{:width=“900px”}
In the figure above:
- General overlays are overlays accessible for every user viewing the content
- User-provided overlays menu becomes available upon login
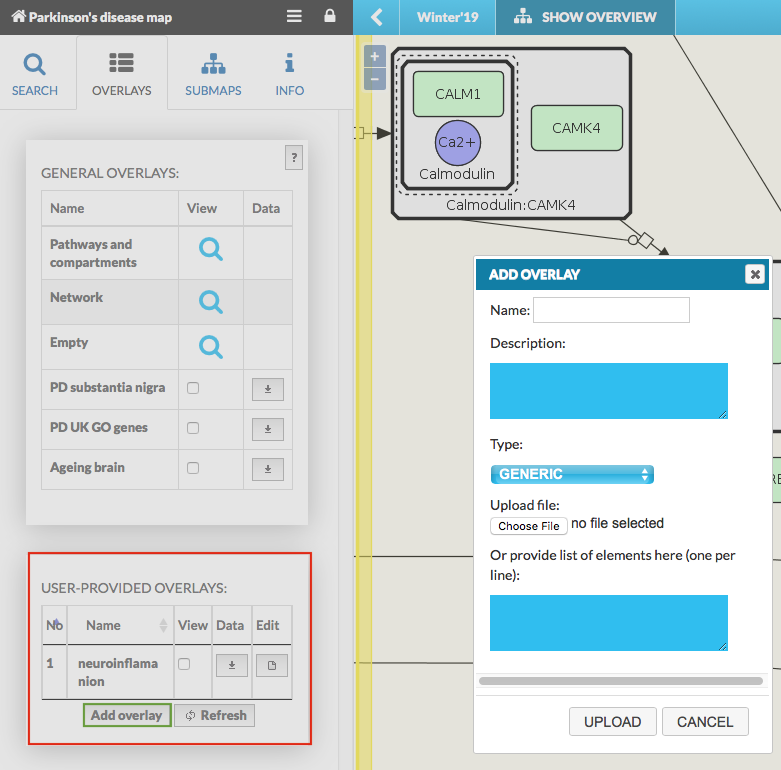
User-provided overlays#
User-provided overlays are the overlays uploaded by and visible to the currently logged user. Available overlays can be reordered by name or number. Hover over any of them display short description, if one was provided on the upload. Adding a new overlay is available via Add overlay button (see below image on the left side), which opens the dialog window (see below image on the right side):
 {:width=“700px”}
{:width=“700px”}
There are two types of overlays the user can provide: generic and genetic variant.
Generic overlay displays a coloring on the map according to data set of indicated molecules (e.g. differentially expressed genes).
Genetic variant overlay displays a coloring for the genes, for which the variants are present. It also enables the genome browser popup, displaying the position of a variant in the sequence of the corresponding gene. To upload and correctly display the genomics information your instance must contain reference genomes, uploaded by the administrator. Also, the Organism id (e.g. 9606) must be provided for the project, and reference genomes uploaded on MINERVA instance must include its type.
The list of user-provided overlays contains columns (image above):
No number of the overlay.
Name of the overlay.
View column with boxes enabling switching between User-provided overlays.
Data column with buttons allowing the user to download the dataset used to generate the overlay. The users have access only to their respective datasets.
Edit column with buttons allowing the user to edit the description to the uploaded datasets.
Add overlay button invoking the dialog window with the following components:
Name field for the new overlay.
Description of the dataset.
Type field, which depends on the header of uploaded file - TYPE field After choosing a file, the dropdown list TYPE switches automatically between GENERIC/GENETIC_VARIANT as Minerva recognizes file’s header. This happens because genetic variant overlays need to have a specific header field.
Upload file - Choose file button to select a file with overlay data.
List of elements field - instead of providing an overlay file, the user can provide a list of named elements to highlight in the map (e.g. list of genes). Choose TYPE corresponding to the content of your list: GENERIC for named elements; GENETIC_VARIANT for the variants.
Upload button to generate the user-define.
(Optional) if a map is displayed using Google Maps API engine, Google Maps API Terms of Use checkbox will appear.
Refresh allows to refresh the overlays list. It is necessary in case an overlay is added via API during viewing a map in a browser.
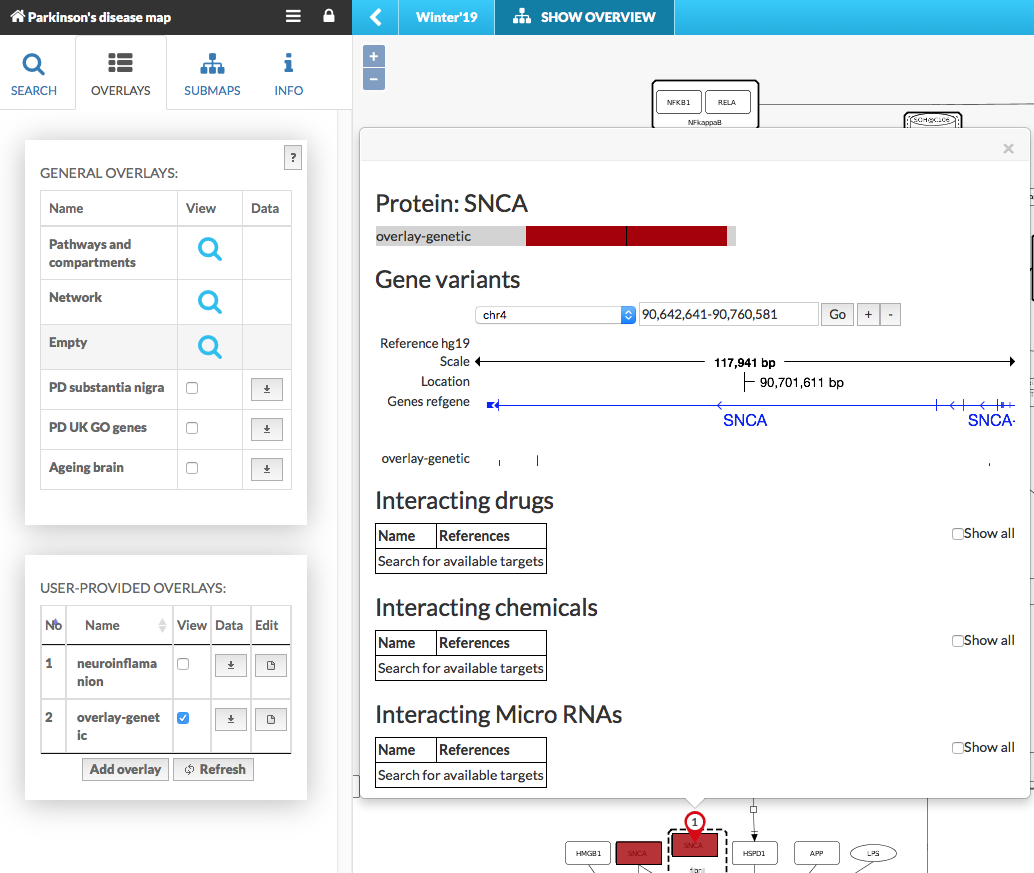
The user-provided overlays are displayed identically as default general overlays. The genetic variant overlays additionally contain the genome browser in the popup window (see below):
 {:width=“900px”}
{:width=“900px”}
The genome browser allows to view the position of a variant in the sequence of the corresponding gene. In Gene variants section (see image above) user can choose the location in chromosome, zoom the scale in/out by clicking on +/- buttons and swipe the base pairs axis. The popup window contain following:
Reference genome contains reference genomes uploaded into the instance by admin (e.g. hg19, hg38)
Scale of base pairs displayed, changed by +/- buttons
Location indicates current position in genome
Genes ref display the reference genes locus provided to the instance by admin, see more in add new genomes
User-provided overlay displays the variants from uploaded file
Upload user-provided overlay data#
Here, we describe how the data overlays files should look like, so MINERVA can handle them and color the map as needed. To learn more about uploading data sets go to example 3 - Custom data upload.
Basic generic format#
The file in a basic format is a two-column, tab-separated text file, with the columns Name and Value. Please note that the basic format does not have a header.
Name column contains the names of elements to be colored
Value column contains the values normalized to [-1, 1] range
Basic format will match names of the elements provided in the Name column among the names of elements in a given network and for the matching ones will assign them colors: blue for negative values, red for positive values, with the saturation proportional to the value. The color range for the data upload is customizable in the admin panel.
EXAMPLE: Click here to download an example of basic format overlay. You can upload it to Parkinson’s disease map.
Advanced generic format#
The advanced format allows for by identifier matching, custom color assignment and coloring of interactions. The uploaded dataset has two sections - the header and the body.
- The header lines start with ‘#’ and can contain the following elements:
- Version -
# VERSION=xyz- a version of this overlay file - Name -
# NAME=xyz- a name that will be automatically assigned upon upload - Description -
# DESCRIPTION=xyz- a description that will be automatically assigned upon upload
- Version -
- The body is a table with a set of allowed columns. See the list of all available columns here.
EXAMPLE: Click here download an example of generic advanced overlay. You can also upload it to Parkinson’s disease map.
The dataset for upload may be integrated and sparse, i.e. a document may contain all columns at once, and, where irrelevant, their content may be left blank. In other words, you can color interactions and elements in the same dataset, leaving blank fields in element_identifier and line_width for elements, and leaving blank fields in name for interactions.
All data columns work as an intersection to narrow down the coloring. Example: if your map contains only protein SNCA, and you put RNA SNCA, there will be no coloring even though the SNCA is displayed in the map.
Genetic variant format#
It is a particular type of advanced format, it contains the header and the body, see below:
- The header lines start with ‘#’ and can contain the following elements:
Type -
#TYPE=GENETIC_VARIANTGenome type -
#GENOME_TYPE=UCSC- type of reference genome being used (e.g. UCSC)Genome version -
#GENOME_VERSION=xyz- version of reference genome being used (e.g. hg19)
- The body is a table with a set of allowed columns. Click here to see the list of all available columns.
EXAMPLE: Click here to download an example of genetic variant overlay, to have an insight on the file’s content. You can also upload it to Parkinson’s disease map.
Upload Choose the dataset to upload and press the Upload button. You will be notified by email when your overlay is ready. The email will also contain the list of elements in your dataset and the number of elements they colored in the map.
Important reminder:
- The number of available overlays is configured in the Administrator’s manual - Users.
- The number of available overlays is common for all projects hosted on your MINERVA instance. If a user is registered in a number of projects and uploads custom datasets in all of them, the global number of available custom overlays will be reduced with each uploaded overlay.
Submaps tab#
The Submaps tab summarizes all the submaps linked to the main map of this project hosted by your MINERVA instance. See Administrator manual - advanced upload to learn how to upload the submaps together with the main file. The tab Submaps is visible in the left panel only if the map contains at least one submap. The tab contains names of the submaps, + to show maps’ description, if one is provided, and the buttons to display them.
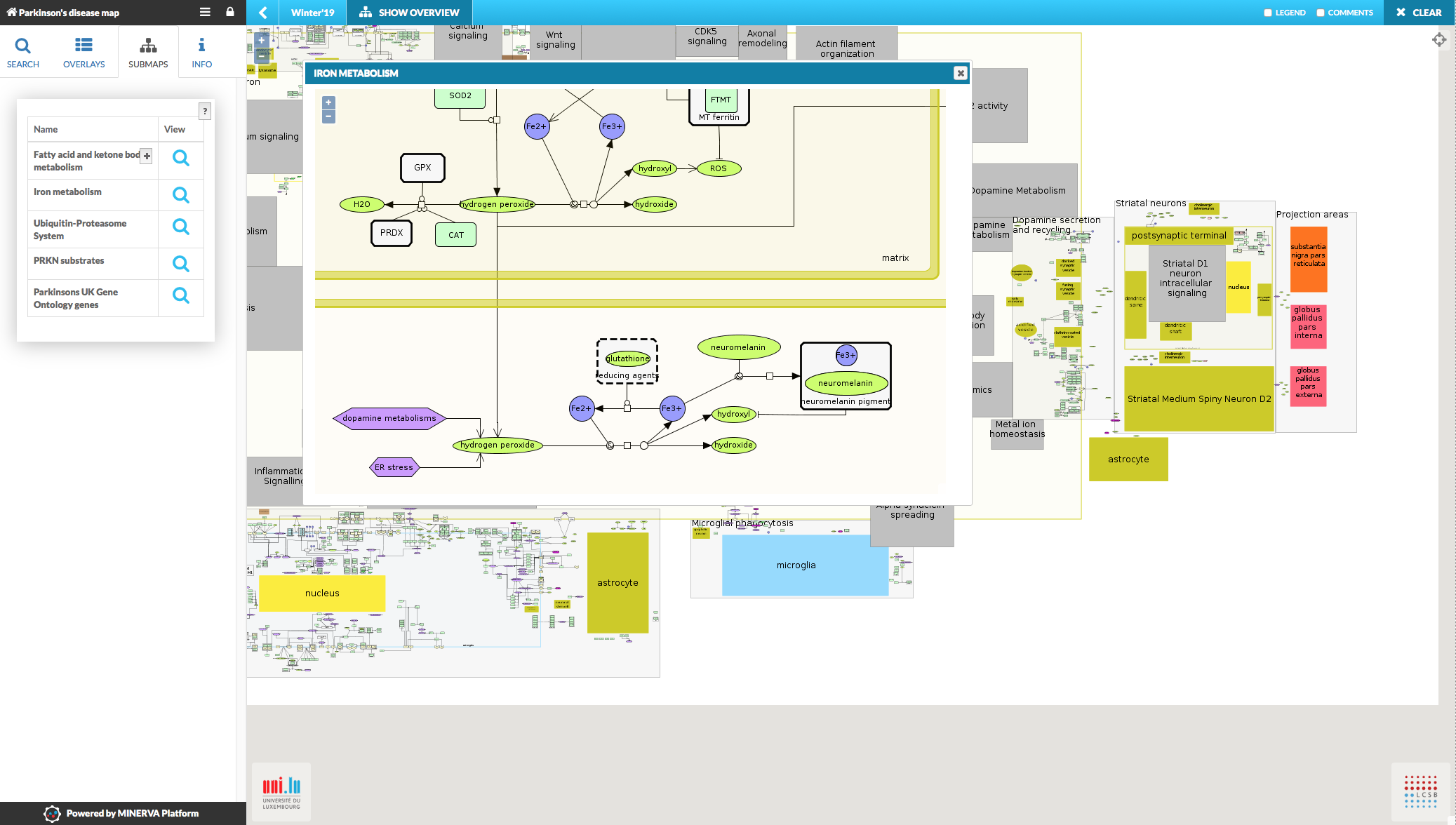 {:width=“900px”}
{:width=“900px”}
The submaps show as a pop-up window on top of the main map, and are synchronized with respect to search queries and displaying data overlays. This means that search results and search pins will be visible in the main map and the active submaps. Similarly, overlays are mirrored in the submaps. Also, on the main map there is an icon informing that search results hit submap. See figures below:
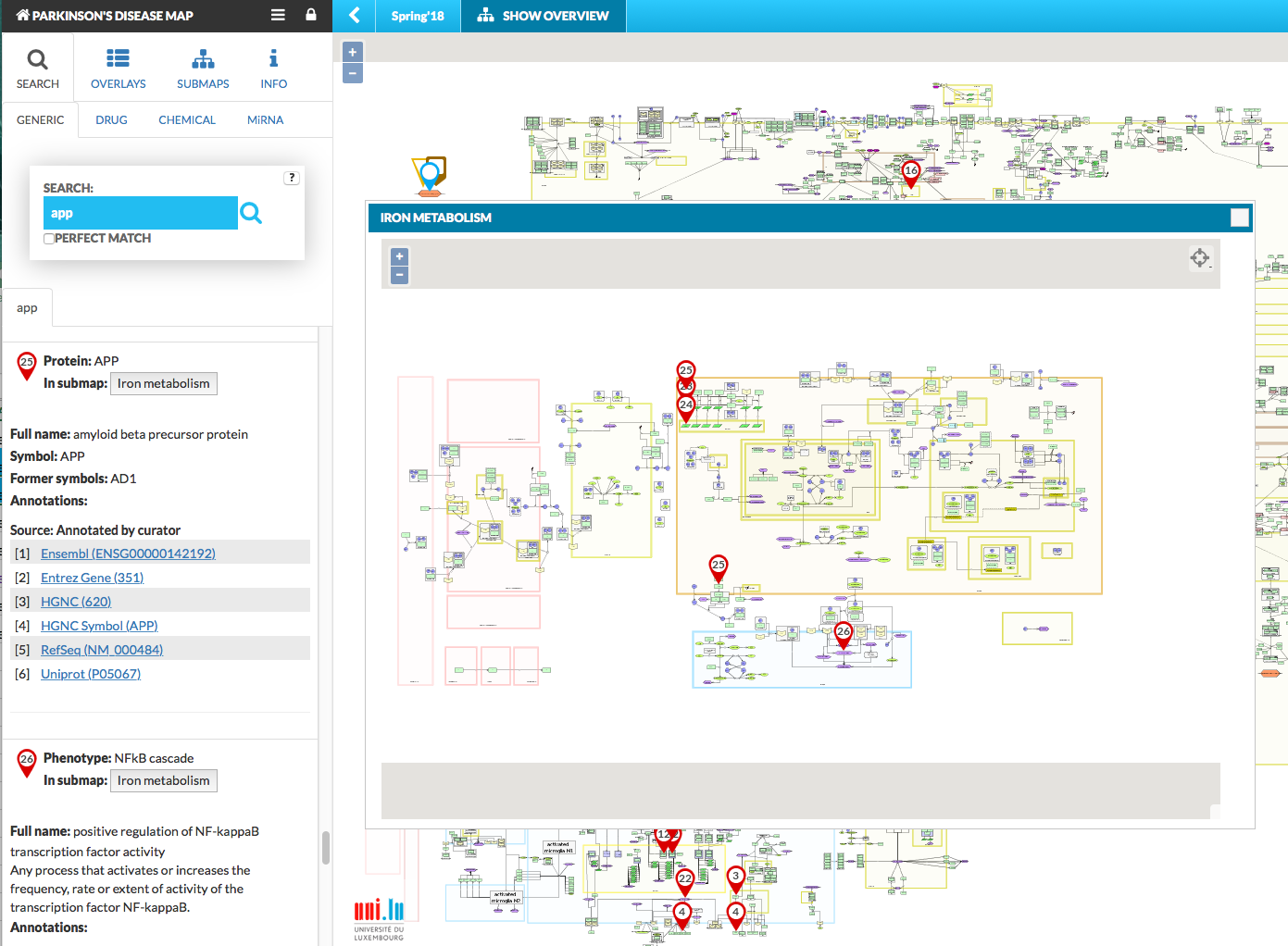 {:width=“900px”}
{:width=“900px”}
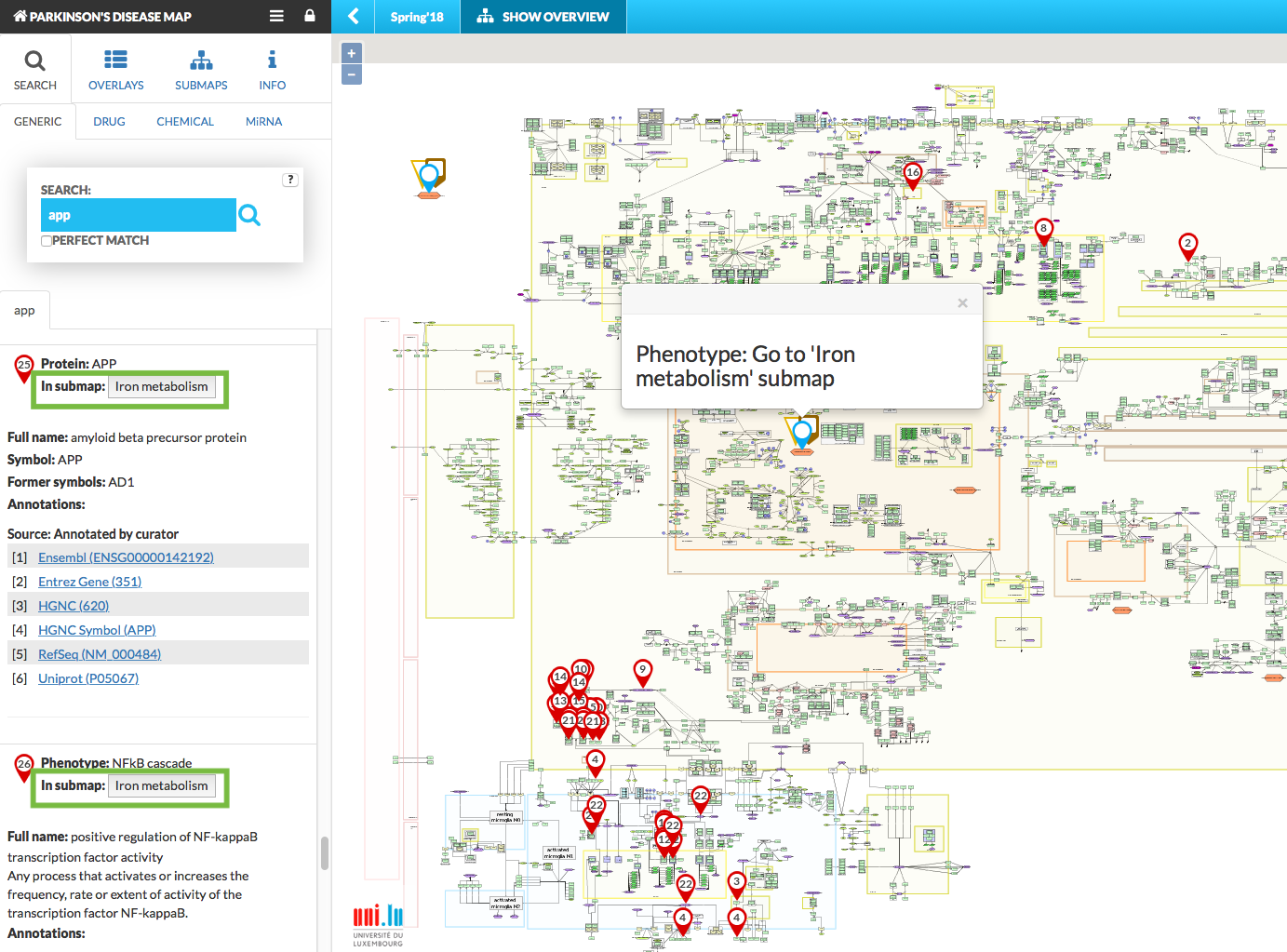 {:width=“900px”}
{:width=“900px”}
Info tab#
The Info tab has two components, displaying Project info and User data (for registered and logged users).
Project info#
Displays name, version and the details of the map.
The Data section features the links to:
- Browse the publications supporting the map. The Publication list dialogue allows to search for articles by title, author names or year. It contains links to interactions supported by particular articles. The list of the publications can be downloaded.
- Download the project’s source files.
- Export the contents of the map (see below for more details).
- Open the MINERVA manual.
- The information about the disease type (Mesh id).
- The information about the organism type (Taxonomy id).
The Description section contains information from the CellDesigner file from Model Notes and Model Description (if provided).
Export#
The Export page contains three tabs (Elements, Network and Graphics), allowing to download the map contents in different formats.
- ELEMENTS tab allows to download the list of elements in the map as a text file. Available filters include:
- Type - filters elements by the element type (e.g. Protein, RNA)
- (sub)map - filters elements by the (sub)map
- Column - selects particular details, including:
- Id - identifier of the element in the MINERVA system
- Name - name of the element
- Map id - identifier of the (sub)network containing this element
- Type - type of element
- Complex id - if applicable, id of the complex containing this element
- Compartment - the smallest compartment/pathway containing the element
- Element external id - corresponding source file e.g. CellDesigner ID
- Annotations - filters the annotations of elements in the downloaded file; by default all annotation types are considered. Fields highlighted in green are non-empty annotation sources
- Included/excluded compartment/pathways - filters the elements by compartments/pathways they are in; by default all compartments are considered as “included”, none as “excluded”
- NETWORK tab allows to narrow down and download the interactions in the map. The exported network file is not a fixed-column table. As interactions are in fact hyper-edges, each line may contain different number of reactants, products and identifiers. Available filters include:
- Element type - filters the nodes of exported interactions by the element type
- Reaction type - filters interactions by type
- (sub)map - filters elements by the (sub)map
- Column - selects particular columns, including:
- Elements - information about reactants, products and reaction modifiers
- Reaction external id - reaction identifier from the source file e.g. CellDesigner ID
- Map id - identifier of the (sub)network containing this element
- Annotations - filters the annotations of elements and interactions in the downloaded file; by default all annotation types are considered. Green highlighted fields are non empty annotation sources
- Included/excluded compartment/pathways - filters the elements by compartments they are in; by default all compartments are considered as “included”, none as “excluded”
- GRAPHICS tab allows to download graphics of the (Sub)map in one of the Format: PNG image, PDF and SVG image.
User data#
If registered, users can login here, or request for an account from the administrators of this MINERVA instance. Platform administrators can register new users (see Administrator’s manual - USERS) and configure Request an account functionality (see Administrator’s manual - Configuration)
After login, this panel displays information about the user. After the successful login the user gains the possibility to upload custom overlays (see Overlays).
INFORMATION BAR#
Information bar is the topmost part of the user view, containing the Show overview button, Comment and Legend checkboxes and Clear button. The Clear button clears all search results currently shown in the display area. The remaining functionalities are detailed below.
Show overview#
This button invokes a static image associated with the displayed content. It may be a graphics helping to understand the underlying map, or any other visual cue that the content curator decided to present. The initial image displayed with the Show overview button can be linked to:
- another static image to be displayed next
- a defined area in the displayed content
- a set of elements or interactions
Comments#
If this checkbox is checked, the comments provided by users with Pinned option set to Yes will become visible in the display area (see Add comment).
Legend#
If this checkbox is checked, the legend describing element and interaction types will be displayed.
PLUGINS#
Plugins allow for creating client-side custom visualizations, independent from the core functionalities of MINERVA. After clicking on the plugin icon a popup window becomes available (see images below).
![]() {:width=“400px”}
{:width=“400px”}
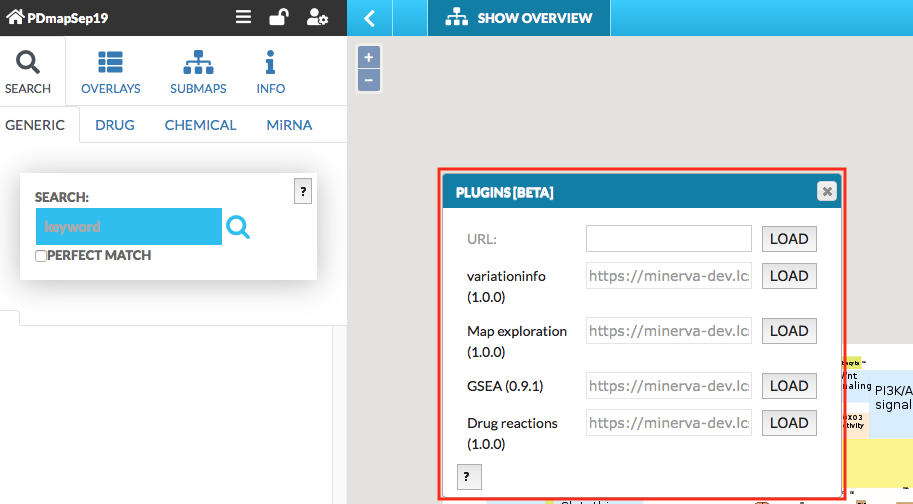 {:width=“600px”}
{:width=“600px”}
The user can choose any of the plugins that are available on the MINERVA platform, uploaded on the instance by the admin. Also, in the URL field one can supply a URL to the plugin, written in JavaScript. To learn how to write your own plugins, visit the “Plugin starter kit” repository.
Example plugin: you can upload below URL into the plugin dialog to find out more:
https://raw.githubusercontent.com/davidhoksza/minerva-plugins-starter-kit/master/dist/plugin.js
Afterwards, a plugin panel becomes available on the right (see image below). To close that panel, click the plugin icon to open the window and click Unload button.
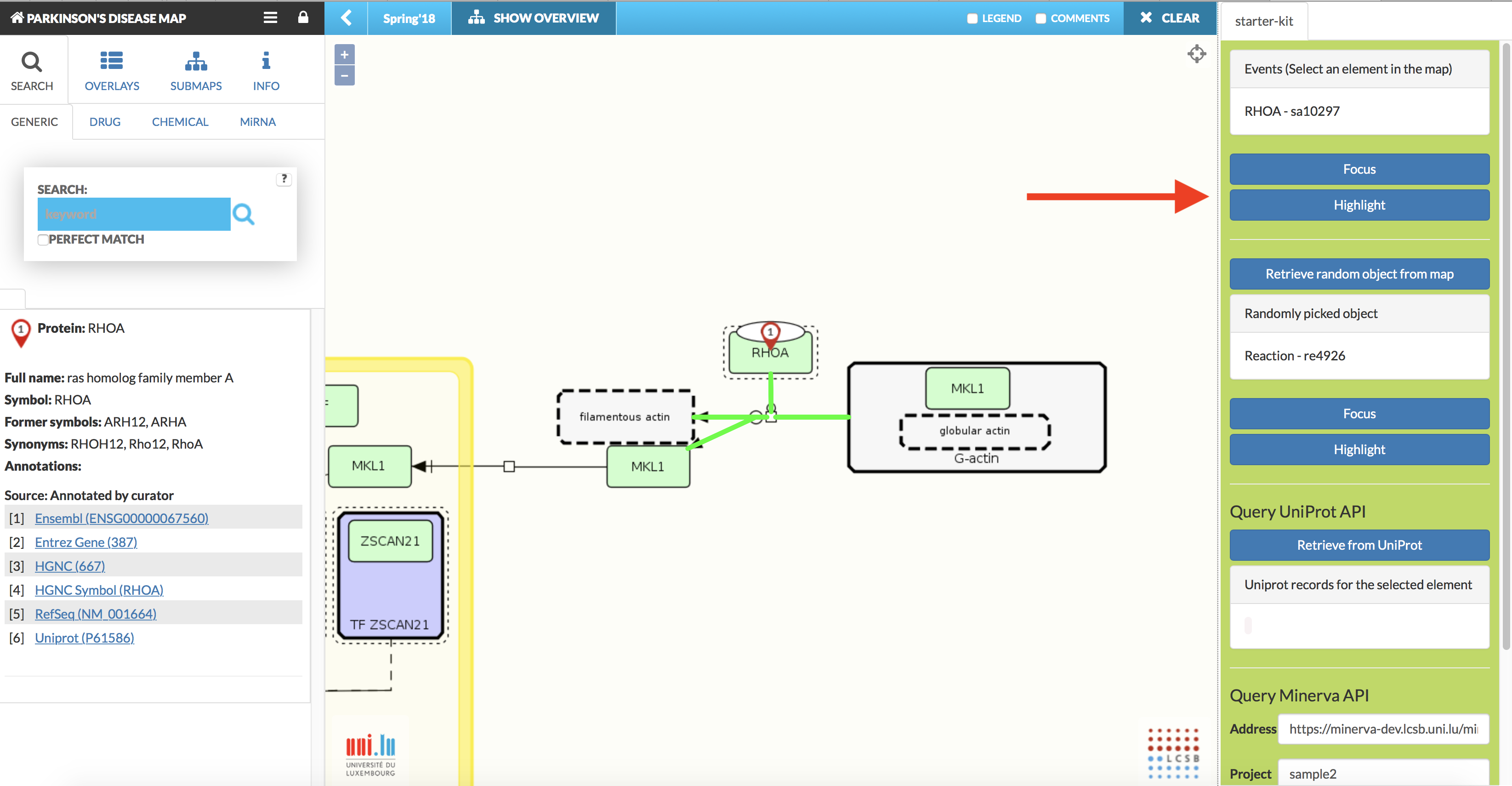 {:width=“900px”}
{:width=“900px”}
Please find below the list of available plugins, click at plugin’s name for its details.
— |
Name of the plugin | Address URL and the short description of the plugin |
Disease variants associations | https://minerva-dev.lcsb.uni.lu/plugins/disease-associations/plugin.js |
| Provide the information about genes associated with the given disease in the context of the selected disease map. |
Drug reactions | https://minerva-dev.lcsb.uni.lu/plugins/drug-reactions/plugin.js |
| Explore the adverse reactions of the drugs which are interacting with the presented entities in a disease map. |
Exploration | https://minerva-dev.lcsb.uni.lu/plugins/exploration/plugin.js |
| Enhance the visualization and exploration of molecular interaction in the map. |
Guide | https://minerva-dev.lcsb.uni.lu/plugins/guide/plugin.js |
| Provide the introductory guide through various aspects of the MINERVA platform. |
GSEA | https://minerva-dev.lcsb.uni.lu/plugins/gsea/plugin.js |
| Calculate the enrichment of the elements from the selected overlay in the pathway. |