Variation exploration#
The variation information plugin allows to see the information on the genetic variations, which are available in public resources in the context of a current disease map.
- table of contents
General instructions#
In order to use the precompiled and publicly available version of the plugin, open the plugin menu in the MINERVA upper left corner and enter below address in the URL field:
https://minerva-dev.lcsb.uni.lu/plugins/variationinfo/plugin.js
The plugin shows up in the plugins panel on the right hand side of the screen.
Plugin functionality#
- The users inputs the name of a disease of interest as a plain string
- The plugin connects to the Proteins API variation service and retrieves all available variants for that disease.
- The returned variants are already grouped by the isoforms of genes involved in. The plugin groups variants in isoforms by gene name and displays them in a table, where each row contains the gene name, number of occurrences of that gene in the map and number of isoforms obtained from the Protein API.
- The user can click on the eye icon of a particular gene to show its occurrences in the map. If there is no occurrence of the given gene in the map, the number of occurrences is zero and no eye icon is displayed.
- By clicking on the plus symbol in each line, the user can explore all the data which was obtained from the Protein API for that gene.
Data resources#
The information about the variants comes from EBI Proteins API. As stated on the pages of Proteins API: >The variation, proteomics and antigen services provide annotations imported and mapped from large scale data sources, such as 1000 Genomes, ExAC (Exome Aggregation Consortium), TCGA (The Cancer Genome Atlas), COSMIC (Catalogue Of Somatic Mutations In Cancer), Peptide Atlas, MaxQB (MaxQuant Database), EPD (Encyclopedia of Proteome Dynamics) and HPA, along with UniProt annotations for these feature types (if applicable)“.
The following image shows an example of the data (a snippet from Google Chrome DevTools) already grouped by gene name and species information (see above).
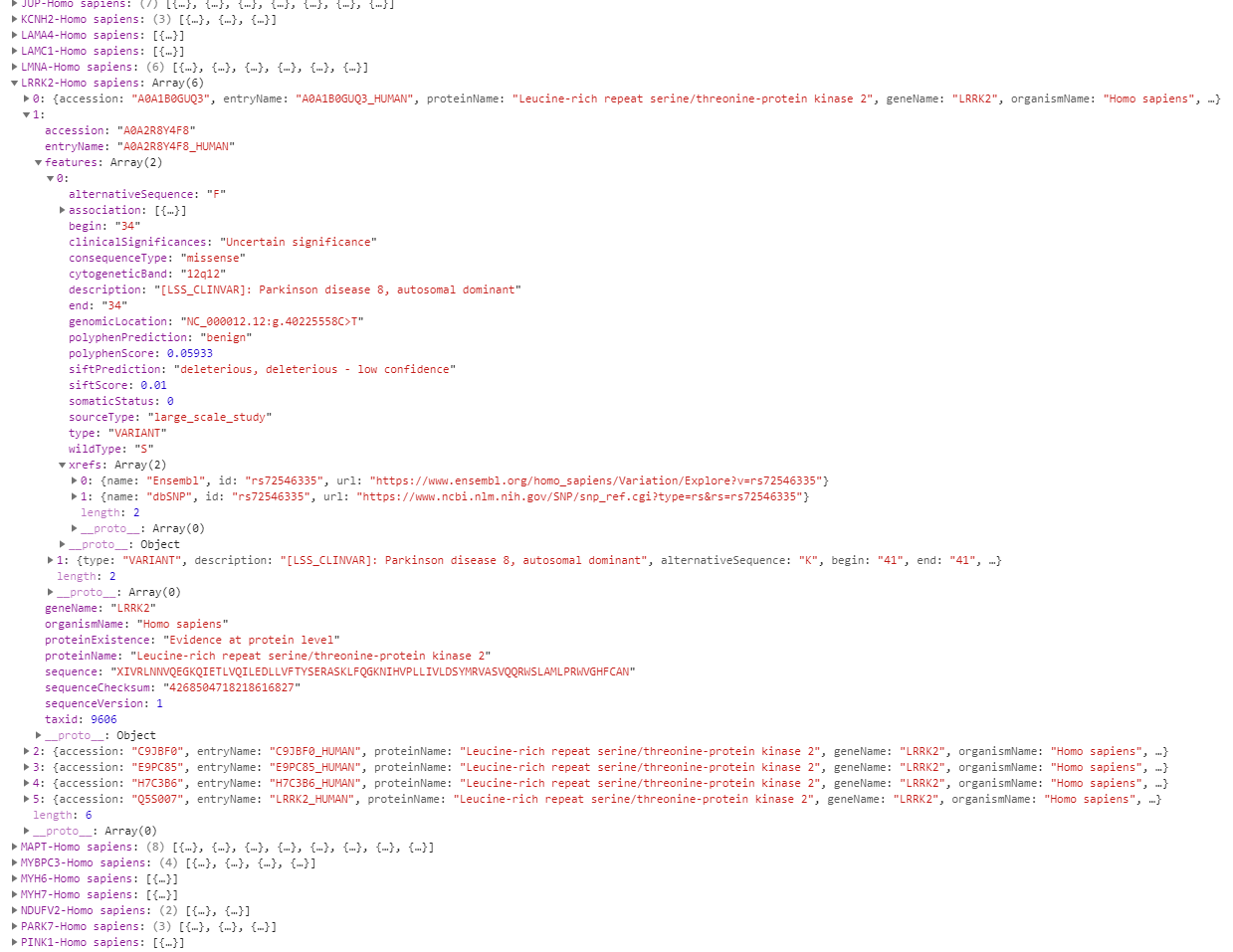 {:width=“900px”}
{:width=“900px”}
Known issues#
The disease name can be entered only as a general string and not as an ontology term. Therefore, queries such as “Parkinson” and “Parkinson’s” will return two distinct sets of variants.