Go back to Examples v12.1
Example 01 - CellDesigner file upload and annotation#
This section provides examples of usage of automatic annotators, automatic verifiers and construction of procedural semantic zoom.
This is Example File 1 containing a CellDesigner file. The file is quite simple, and has the following properties:
- Annotations
- Gene component (SNCA) is not annotated
- RNA component (SNCA) is annotated with Entrez Gene id (NCBI Gene in CellDesigner MIRIAM tab)
- one protein component (SNCA) is annotated with UniProt ID, while the other (PRKN) is not annotated
- Phenotype component (UPS) is annotated with Gene Ontology
- one interaction re3 is annotated wit PubMed id, while others are not
- Compartments and visual layers
- Translation and Degradation visual layers are drawn on top of the diagram
- cell, synapse and nucleus compartments are drawn
Automatic annotation#
See Configure automatic annotation for details on annotators.
- Upload the Example File 1 to your MINERVA instance using Add project button
- Configure automatic annotations by:
- checking the checkbox Annotate model automatically
- pressing the Advanced button; set the following:
- for Element > Species > Gene, set HGNC annotator
- for Element > Species > Phenotype, set Gene Ontology annotator
- for Element > Species > Rna, set Entrez annotator
- for Element > Species > Protein > GenericProtein, set UniProt annotator
- Press Submit
Examine the generated content. Please note that:
- Annotations between SNCA DNA, RNA and Protein are different, because different annotators used different information sources to obtain the information on the elements. Annotations with source: Annotated by curator are marked separately from all results (these annotations mirror MIRIAM annotations within source file)
- Even though SNCA DNA had no CellDesigner annotation, it was correctly annotated by name by HGNC annotator
- UniProt annotator works only by identifier, so it was unable to provide annotations for PRKN (it is not annotated in CellDesigner)
- Gene Ontology annotator retrieved a definition of the annotated Phenotype
Automatic verification#
See Section Configure automatic verification for details on annotators.
NOTE: Your MINERVA instance remembers the settings from the previous run, so keep annotation settings as above.
- Upload the Example File 1 to your MINERVA instance again. Make sure to either remove previously uploaded example, or to name the uploaded project differently (use Add project button)
- Configure automatic verification by:
- checking the checkbox Verify manual annotations
- pressing the Advanced button and set the following for:
- Element > Species > Gene
- select as valid (top right window): HGNC, HGNC Symbol, Entrez Gene, Ensembl
- tick checkbox One of these miriam should be required and set as required (bottom right window): HGNC, HGNC Symbol, Entrez Gene, Ensembl
- Element > Species > Phenotype
- select as valid: Gene Ontology
- tick checkbox and set as required: Gene Ontology
- Element > Species > Rna
- select as valid: HGNC, HGNC Symbol, Entrez Gene
- tick checkbox and set as required: HGNC, HGNC Symbol and Entrez Gene
- Element > Species > Protein > GenericProtein
- select as valid: UniProt, HGNC Symbol, Entrez Gene
- tick checkbox and set as required: UniProt
- GenericReaction > StateTransition
- select as valid: PubMed
- tick checkbox set as required: PubMed
- GenericReaction > Translation
- select as valid: PubMed
- uncheck One of these miriam should be required checkbox
- GenericReaction > Transcription
- select as valid: PubMed
- uncheck One of these miriam should be required checkbox
- Make sure no items in upper hierarchy levels (e.g. Element, Species or Protein for GenericProtein) have different sets of verification rules
- Press Submit
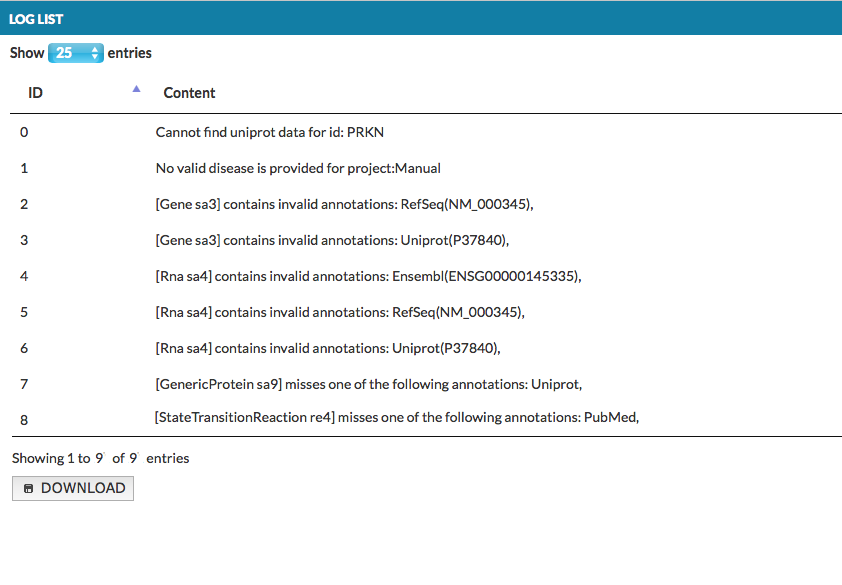
Upon upload, an exclamation mark icon will appear next to the project. After clicking, the popup window will look like this:
 {:width=“900px”}
{:width=“900px”}
Examine the generated warnings. According to the above image, please note that:
- PRKN is missing UniProt ID in CellDesigner
- Automatic annotators provided additional annotations to Gene and RNA elements, outside of the valid list, you can suppress these warnings by updating the valid list
- GenericProtein element (PRKN) was indicated as missing a required annotation (UniProt)
- StateTransition re4 was indicated as missing a required annotation (PubMed) See more in section Examine warnings
- TranslationReaction and TranscriptionReaction did not raise any warnings as no annotation was required for them
NOTE: Valid and Required settings for lower hierarchy levels are overridden by settings for upper hierarchy levels. For instance, if no PubMed annotation is required for TranslationReaction, but Reaction requires PubMed, reactions re1 and re2 raise a warning.
Procedural semantic zoom#
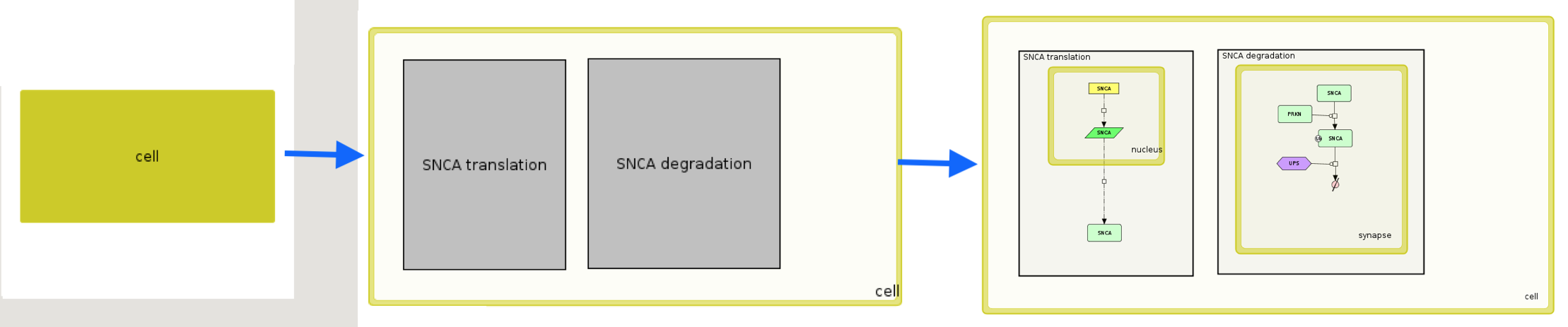
Example File 1 after upload and submit will generate a view with procedurally-generated semantic zoom. Please, note that on the highest zoom out level only one compartment - cell is visible. After zoom-in, two grey areas become visible, describing functional areas in the map. After another zoom-in, nucleus and synapse compartment become visible. See also Add project button.
 {:width=“900px”}
{:width=“900px”}